How Experiments Evolve
The phylomethodic tree
At Briefly, we often use code as an analogy for scientific methods. Where code is a set of instructions that a human writes for a computer to run, a method is a set of instructions that a scientist physically executes in the lab.
So how does great software get built? Is there anything we can learn as scientists to help us develop and maintain great methods and spend less time troubleshooting? We think version control – a key tool in grappling with code as it grows and evolves, could be particularly powerful if appropriately applied to scientific experiments.
Version control in software development
Let’s look at a simple example in software. A developer has built and released a janky version of their app. It gets the job done, but it’s slow, so they start working on speeding it up. They iterate on the code for a little while, making some tweaks here and there. When they come to run their new version, ah crap, these changes break the whole app, and worse, our software engineer has no idea why. This is not a reason to panic. Here’s why:
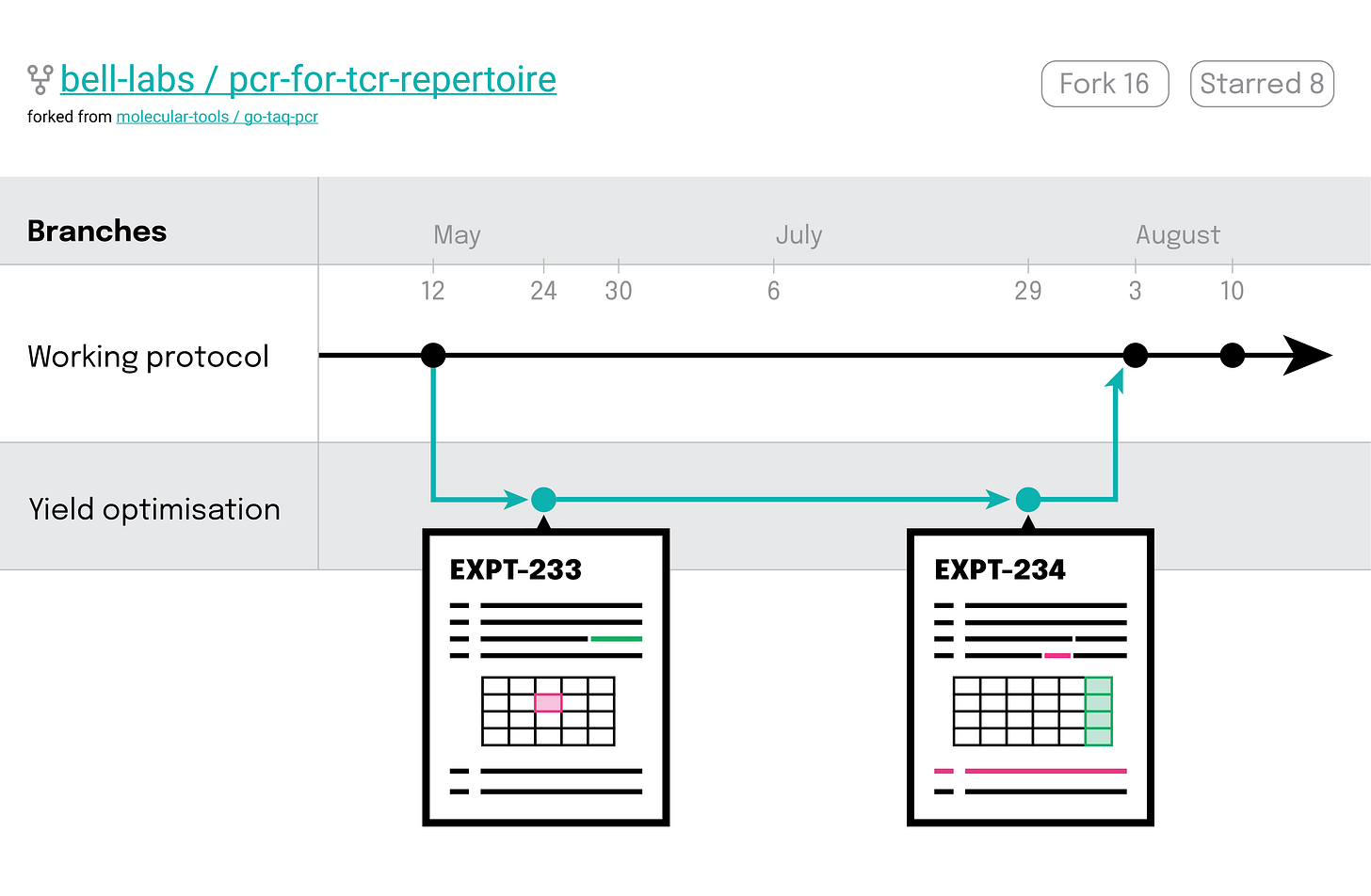
All of this development work is happening separately from the working app, on a ‘branch’ – the old, slow, janky version may not be perfect, but it works and they can always get back there.
As our developer has been writing this code, they have been registering their changes (‘commits’) as they go along. Their version control system allows them to trace back through these changes, seeing how their code evolved during development, helping them figure out what may have broken it. If they’re stuck, they can always just revert back to a previous version and start again from there.
Bringing version control to science
To us, some kind of version control has always felt like a natural fit for scientific experiments. We study complex interacting systems, where small changes in our methods can result in our experiments going awry in unexpected ways. Our experiments & methods also have a lineage, with each new design based on previous work. Version control could:
Help us separate development work from working methods. By developing or optimising methods on a branch, we can protect our most precious samples from unvalidated protocols.
Enable us to explore the lineage of our experiments - to see the evolution of each and every parameter and step and to understand how and why changes occurred.
In short, it would give us the ability to time travel through our previous experiments. We could spend less time leafing through countless notebook entries to figure out where things went wrong. Or how exactly we got that interesting result a few months back.
What’s stopping us?
When a developer looks back at a previous version of their software, they get the entire codebase as it was at that point in time. They can see each and every change that has occurred since then, and can piece together what might have gone wrong.
Scientists on the other hand have a lab notebook entry. As I discussed in a previous post, these are unstructured and rarely contain every detail of an experiment – documenting in this way is just an unrealistic amount of work.
The result is that many mutations in experimental design become invisible, and accumulate over time. They exist in a single lab notebook entry and in the head of the scientist. Some may be passed down from senior scientists to juniors. But there is no phylogeny – no record of where and why those changes first appeared, how old datasets were generated in the past, or whether certain tweaks are still relevant in the experiments we’re running now.
Moving away from the notebook
To enable scientists to track how their experiments change over time, we need to move away from the free text lab notebook as the primary source of truth for tracking how our experiments were done. In its place, we need something that
Facilitates and actively helps scientists to completely capture the details of each of their experiments.
Captures these details with a more consistent format and language, such that experiments can be compared directly, back-to-back.
Tracks these changes actively over time, experiment-by-experiment, so that scientists can revisit the full picture of an experiment at any point in the future.
How we get there
Again, we can look to software engineering for some inspiration. We have countless tools that actively help engineers write good quality code. One basic example is the code linter. Linters analyse and flag issues while you’re coding. This happens automatically, in the background. Oops you forgot to define this variable, close that brace, or flag that you’re writing in an inconsistent style.
Today, AI-enabled tools like Github Copilot are taking things a few steps further – not just spotting semantic issues with what the developer has written, but actively drafting code for them based on their ideas and requests.
We’re building something similar for scientists – a tool that takes scientists' ideas and works with them to create clear, consistent, complete experimental designs, spotting details that are missing (‘what CO2% are you growing these cells in?’), and suggesting potential issues and pitfalls (‘You’re using the wrong antibiotic for that plasmid!’).
We think a tool like this could help us reach the consistent, complete experimental documentation I described earlier. With it, we can unlock version control for scientific experiments, tracking changes over time, and building up the phylogeny behind each method and experimental design.
Interested? We're looking for scientists who are searching for a better way to organise and track their experiments. We want to build this with you!